Drawing Classes
In VARNA API, all drawing classes are inherited from the class BasicDraw. We offer three Python classes Structure, Comparison, and Motif for different RNA secondary structure visualisation. The first two correspond to the classic and the comparison mode in VARNA. The last one is the special case for motif drawing.
- Structure: The most common drawing class taking a secondary structure and/or an RNA sequence as input.
- Comparison: Comparison between two structures and two sequences.
- Motif: Customized class to draw motif, an union of loops.
BasicDraw
Bases: _VarnaConfig
Source code in src/varnaapi/models.py
83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269 270 271 272 273 274 275 276 277 278 279 280 281 282 283 284 285 286 287 288 289 290 291 292 293 294 295 296 297 298 299 300 301 302 303 304 305 306 307 308 309 310 311 312 313 314 315 316 317 318 319 320 321 322 323 324 325 326 327 328 329 330 331 332 333 334 335 336 337 338 339 340 341 342 343 344 345 346 347 348 349 350 351 352 353 354 355 356 | |
add_annotation(annotation)
Add an annotation. Argument should be a valid Annotation object
Examples:
>>> v = varnaapi.Structure()
>>> a = varnaapi.param.LoopAnnotation("L1", 6, color="#FF00FF")
>>> v.add_annotation(a)
Source code in src/varnaapi/models.py
159 160 161 162 163 164 165 166 167 168 169 170 171 | |
add_aux_BP(i, j, edge5='wc', edge3='wc', stericity='cis', color='blue', thickness=1, **kwargs)
Add an additional base pair (i,j), possibly defining and using custom style
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
i |
int
|
5' position of base pair |
required |
j |
int
|
3' position of base pair |
required |
edge5 |
str
|
Edge 5' used for interaction in non-canonical base-pairs, as defined by the Leontis/Westhof classification of base-pairs. Admissible values are wc (Watson/Crick edge), h (Hoogsteen edge) and s (Sugar edge). |
'wc'
|
edge3 |
str
|
Edge 3' used for interaction in non-canonical base-pairs. Admissible values are wc, h and s. |
'wc'
|
stericity |
str
|
Orientation of the strands. Admissible values are cis and trans |
'cis'
|
color |
color
|
Base-pair color |
'blue'
|
thickness |
float
|
Base-pair thickness |
1
|
Source code in src/varnaapi/models.py
99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 | |
add_bases_style(style, bases)
Apply a BasesStyle to a list of positions. If a position is assigned to more than one styles, one of them will be randomly used.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
style |
BasesStyle
|
Style to apply |
required |
bases |
list
|
List of 0-indexed positions |
required |
Examples:
>>> v = varnaapi.Structure()
>>> style1 = varnaapi.param.BasesStyle(fill="#FF0000")
>>> style2 = varnaapi.param.BasesStyle(fill="#FFFF00" outline="#00FF00")
>>> v.add_bases_style(style1, [1,2,4])
>>> v.add_bases_style(setye1, [10,11,12])
>>> v.add_bases_style(style2, [4,5,6,7])
Source code in src/varnaapi/models.py
136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 | |
add_chem_prob(base, glyph='arrow', dir='in', intensity=1, color='#0000B2', **kwargs)
Add chemical probing annotation on two adjacent bases.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
base |
int
|
index of the first base of adjacent bases |
required |
glyph |
str
|
Shape of the annotation chosen from ['arrow', 'dot', 'pin', 'triangle'] |
'arrow'
|
dir |
str
|
Direction of the annotation chosen from ['in', 'out'] |
'in'
|
intensity |
float
|
Annotation intensity, i.e. thickness |
1
|
color |
color
|
Color used to draw the annotation |
'#0000B2'
|
Source code in src/varnaapi/models.py
173 174 175 176 177 178 179 180 181 182 183 184 | |
add_colormap(values, vMin=None, vMax=None, caption='', style='energy', **kwargs)
Add color map on bases.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
values |
float list
|
list of values in float for each base. |
required |
vMin |
float
|
Minium value for the color map |
None
|
vMax |
float
|
Maximum value for the color map |
None
|
caption |
str
|
Color map caption |
''
|
style |
Color map style, which is one of the following
|
'energy'
|
Source code in src/varnaapi/models.py
186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 | |
add_highlight_region(i, j, radius=16, fill='#BCFFDD', outline='#6ED86E', **kwargs)
Highlights a region by drawing a polygon of predefined radius,
fill color and outline color around it.
A region consists in an interval from base i to base j.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
i |
int
|
5'-end of the highlight |
required |
j |
int
|
3'-end of the highlight |
required |
radius |
float
|
Thickness of the highlight |
16
|
fill |
color
|
The color used to fill the highlight |
'#BCFFDD'
|
outline |
color
|
The color used to draw the line around the highlight |
'#6ED86E'
|
Source code in src/varnaapi/models.py
115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 | |
dump_param(filename)
Store style parameters into file
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
filename |
path to the file |
required |
Source code in src/varnaapi/param.py
627 628 629 630 631 632 633 634 | |
enable_smart_flip(enable=True)
Enable to flip positions treating to address points 1-3 in flip(). When enable, for each branch of exterior loop, VARNA API will send only the 5' most position to flip to VARNA if any position (unpaired included) of the branch is given by flip().
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
enable |
bool
|
Enable or disable smart flip. |
True
|
See Also: BasicDraw.flip
Source code in src/varnaapi/models.py
236 237 238 239 240 241 242 243 244 245 | |
flip(*positions)
Flip one or more helices identfied by given positions.
Behind the flip
For a given base or basepair, VARNA flips the helix the base or the basepair belongs to by identifying first the farest position at 5' and then redrawing the helix in the counter direction from that position. By default, VARNA positions bases in clockwise direction, therefore repositioning bases in counter clockwise direction gives the effect of flip. Such flipping rule gives the following results:
1. No flip will happen if given position is unpaired.
2. Giving even number of positions of the same helix cancels out the flip.
3. Consider two helices separated by a loop. Giving the position of the first helix flips both helices as one. However, giving the position of the second helix will result the flipping of only the second one, which may cause two helices overlap in the drawing.
4. In linear drawing mode, flipping will not draw basepair arcs in lower plane as if affects bases positioning.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
positions |
either a base in integer or a basepair in integer tuple of the helix to flip |
()
|
Examples:
Consider secondary structure
...(((...)))...((...))...(((...)))...
1234567890123456789012345678901234567
>>> v = varnaapi.Structure(structure=dbn)
>>> v.flip(5, (27,33))
See Also: BasicDraw.enable_smart_flip
Source code in src/varnaapi/models.py
204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 | |
get_params(complete=False)
Get parameters with value in dictionary By default, only the parameters with value different than the default are returned. Set complete to True to get complete parameters.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
complete |
bool
|
Return complete parameters. |
False
|
Source code in src/varnaapi/param.py
576 577 578 579 580 581 582 583 584 585 586 587 588 | |
load_param(filename)
Load existing style parameters from file
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
filename |
path to the file |
required |
Source code in src/varnaapi/param.py
636 637 638 639 640 641 642 643 | |
savefig(output, show=False)
Call VARNA to draw and store the paint in output
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
output |
Output file name with extension is either png or svg |
required | |
show |
bool
|
Show the drawing |
False
|
Note
The argument show is used only in jupyter notebook
Source code in src/varnaapi/models.py
328 329 330 331 332 333 334 335 336 337 338 339 340 341 342 343 344 345 346 347 348 349 | |
set_algorithm(algo)
Set algorithm other than radiate to draw secondary structure. Supported options are line, circular, radiate and naview.
Source code in src/varnaapi/param.py
592 593 594 595 596 | |
set_bp_style(style)
Set default style for base-pairs rendering, chosen among BP_STYLES
Note: lw is set by default
Examples:
>>> v = varnaapi.Structure()
>>> v.set_bp_style("simple")
Source code in src/varnaapi/param.py
598 599 600 601 602 603 604 605 606 607 | |
set_title(title, color='#000000', size=19, **kwargs)
Set title displayed at the bottom of the panel with color and font size
Source code in src/varnaapi/models.py
131 132 133 134 | |
show(extension='png')
Show the drawing
Equivalent to savefig(tmp, show=True) where tmp is a temporary file
Source code in src/varnaapi/models.py
351 352 353 354 355 356 | |
update(loaded=False, **kwargs)
Easy way to update params value. The list of keyward arguments can be found here
Source code in src/varnaapi/param.py
554 555 556 557 558 559 560 561 562 563 564 565 566 567 568 569 570 571 572 573 574 | |
Structure
Bases: BasicDraw
Classic VARNA drawing mode. Constructor from given RNA sequence or/and secondary structure.
If sequence and structure have different size, the larger one is used
and s or .s will be added to sequence or structure to complement.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
sequence |
Raw nucleotide sequence for the displayed RNA.
Each base must be encoded in a single character.
Letters others than |
None
|
|
structure |
str or list
|
RNA (pseudoknotted) secondary structure in dbn |
None
|
Source code in src/varnaapi/models.py
358 359 360 361 362 363 364 365 366 367 368 369 370 371 372 373 374 375 376 377 378 379 380 381 382 383 384 385 386 387 388 389 390 391 392 393 394 395 396 397 398 399 400 401 402 403 404 405 406 407 408 409 410 411 412 413 414 415 416 417 418 419 420 421 422 423 424 425 426 427 | |
Comparison
Bases: BasicDraw
Drawing of two aligned RNAs.
Unlike classic Structure mode,
both sequences and structures MUST be specified and have the same size.
Additionally, the merged secondary structures must currently be without any crossing
interactions (e.g. pseudoknots), and merging them should give a secondary structure.
Gap character is ..
Args:
seq1 (str): Sets the gapped nucleotide sequence for the first RNA sequence
structure1 (str): Sets the first secondary structure in Dot-Bracket Notation
seq2 (str): Sets the gapped nucleotide sequence for the second sequence
strcuture2 (str): Sets the second secondary structure in Doc-Bracket Notation
Source code in src/varnaapi/models.py
435 436 437 438 439 440 441 442 443 444 445 446 447 448 449 450 451 452 453 454 455 456 457 458 459 460 461 462 463 464 | |
Motif
Bases: BasicDraw
Special class for motif drawing.
A motif is a rooted ordered tree, similar to a secondary structure,
but whose leaves may represent base paired positions, named open base
pairs or open paired leaves and denoted by (*), and the root always
represents a closing base pair. A motif can also be seen as an union
of consecutive loops. The figure below represents ((*)(*)(((*)(*)))).
Motif class inherits from Structure with some pre-set parameters.
- rotation is set at
180 - default base pair style is
simple - base number is hidden by setting default color to white (default background color)
A dummy base pair is added after each open base pair and in front of
the root, as shown in the figure below.
Therefore, the index of bases is changed after creating the object.
For example, the index of first base of root is 1 instead of 0.
The default bases style for root is
BasesStyle(fill="#606060", outline="#FFFFFF", number="#FFFFFF") and
BasesStyle(fill="#DDDDDD", outline="#FFFFFF", number="#FFFFFF") for
dummy bases. One can change them using
set_root_bases_style and
set_dummy_bases_style.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
motif |
str
|
Motif in Dot-Bracket Notation.
|
required |
sequence |
str
|
Chain of characters for motif. Note that sequence
should exactly match with motif, i.e. Equal length and same
positions for all |
None
|
Source code in src/varnaapi/models.py
467 468 469 470 471 472 473 474 475 476 477 478 479 480 481 482 483 484 485 486 487 488 489 490 491 492 493 494 495 496 497 498 499 500 501 502 503 504 505 506 507 508 509 510 511 512 513 514 515 516 517 518 519 520 521 522 523 524 525 526 527 528 529 530 531 532 533 534 535 536 537 538 539 540 541 542 543 544 545 546 547 548 549 550 551 552 553 554 555 556 557 558 559 560 561 562 563 564 565 566 567 | |
set_dummy_bases_style(style)
Set style for dummy bases. Argument is a BasesStyle object.
Source code in src/varnaapi/models.py
544 545 546 547 548 549 | |
set_root_bases_style(style)
Set style for root bases. Argument is a BasesStyle object.
Source code in src/varnaapi/models.py
551 552 553 554 555 556 | |
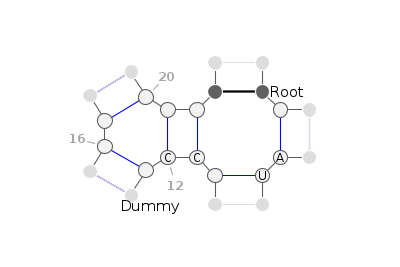
Example
Figure above is created with
from varnaapi import Motif
from varnaapi.param import BaseAnnotation
m = Motif("((*)(*)(((*)(*))))", sequence=" *AU* CC * * ")
m.add_annotation(BaseAnnotation(" Root", 2))
m.add_annotation(BaseAnnotation("Dummy", 14))
# Show how base indices work for motif.
# Remeber that VARNA is 1-indexed
m.set_default_color(baseNum="#a6a6a6")
m.set_numeric_params(periodNum=4)
m.savefig("motif_ex.png")